Making sense of missense in a rare children’s neurological disease
Rare genetic diseases, as the name suggests, are rare. Even diseases that are part of Finnish national heritage, such as aspartylglucosaminuria (AGU), with high prevalence ratios, are diagnosed in only a few children annually. Collectively, however, this “rarity” is an illusion, with an estimated 6,000 to 10,000 different rare diseases affecting up to 8% of the world’s population. Essentially, this phenomenon impacts us all, either directly or indirectly.
Although challenging, rare diseases can be treated similarly to infectious diseases. For example, drugs like elexacaftor and ivacaftor have been developed to alleviate the symptoms of cystic fibrosis. However, because drug development is very expensive, early-stage research, which could lead to breakthroughs, relies entirely on open academic efforts that face their own funding challenges. Regardless, it is morally imperative that we try to tackle this issue, as it affects foremost the most vulnerable and cherished members of our society—children.
SynGAP1-related neurodevelopmental disorders
SynGAP1-related neurodevelopmental disorders are an example of a rare disease in urgent need of our attention. Diagnosis of this disease, manifesting in early childhood, is challenging due to its diverse symptoms including epilepsy, autism, and intellectual disabilities. Despite its rarity (6:100,000 prevalence), it may cause 2–8% of all intellectual disabilities worldwide. The SynGAP1 protein is crucial for synapse development and plasticity, impacting learning and potentially cancer or Alzheimer’s disease development.
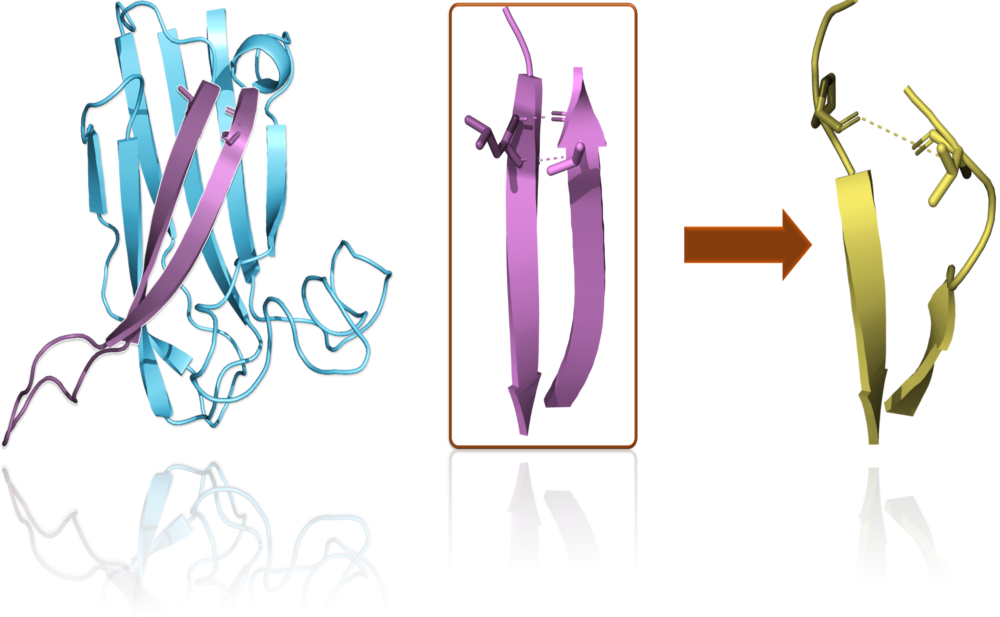
Most SynGAP1 cases are likely due to missense mutations, where one amino acid is replaced by another. These swaps can affect protein folding and structure, making it hard to assess their pathogenicity through sequencing alone. Indeed, our modelling research, focusing on methods like molecular dynamics simulations and folding stability calculations, shows that structure-based approaches provide the best accuracy for assigning pathogenicity to SynGAP1 missense variants.
SynGAP missense server: sharing everything with everybody all at once
Funded by the SynGAP Research Fund (SRF) since 2023, our work emphasizes practicality and openness. We have created the SynGAP Missense (SGM) server (https://syngapmissenseserver.utu.fi) to share modeling results. The SGM server allows users to search for computational predictions and explore SynGAP1 missense variants structurally using a 3D Viewer.
While the “publish or perish” rule certainly looms over us, this project has a refreshing clarity on what counts as a success. If we succeed in giving clarity to families on the cause of their children’s symptoms and make progress, however small, towards effective treatment, we are on the right track.
Our research is conducted in close collaboration with Docent Michael Courtney and Dr. Li-Li Li (Neuronal Signalling Laboratory), Docent Ilkka Paatero (Zebrafish facility), and Dr. Jukka Lehtonen (Structural Bioinformatics Laboratory) at the Turku Science Park. Our common goal is to understand the impact of missense mutations on SynGAP1-related neurodevelopmental disorders and develop new methods for disease diagnosis and drug screening. More broadly, the innovations produced by the research are to be applied to other rare neurological diseases as well.

Pekka Postila
PhD, Docent, Senior Researcher
